Enzymes
Many different reactions take place in biological systems. Experiments have shown that the rate of these reactions when performed outside a living system (ie in vitro ) is much lower than in vivo . In living systems, reactions occur hundreds to millions of times faster than in vitro . This is caused by specific catalysts - enzymes . These often enable the course of such reactions that would otherwise practically not take place in the conditions of the human body (temperature, pH, etc.).
Biological catalysts[edit | edit source]
A catalyst is a substance that increases the rate of a chemical reaction, but does not change the chemical equilibrium (only shortens the time it takes to achieve it). During the reactions, the enzyme molecule is not consumed.
Enzyme[edit | edit source]
An enzyme is a specific organic molecule that accelerates reactions in organisms (it acts as a biocatalyst). This enables reactions to take place even at relatively low temperatures, neutral pH and atmospheric pressure, which are normally found in organisms. The vast majority of enzymes are proteins . The exception is some types of RNA molecules - so-called ribozymes.
In addition to the protein component, enzymes can also contain a non-protein component . According to its presence, enzymes can be divided into:
- simple :
- composite :
- in addition to the protein part (the so-called apoenzyme , it is ineffective by itself), they also contain a non-protein part - the so-called cofactor . The cofactor together with the apoenzyme forms an active enzyme molecule, the so-called holoenzyme.
Cofactors[edit | edit source]
A cofactor can be:
- metal ions: Zn 2+ (e.g. alcohol dehydrogenase), Mn2+ (e.g. arginase), Fe2+ , C 2+ , Mg2+ ,
- organic molecules: they are often derivatives of vitamins.
According to the nature of the binding to the apoenzyme, cofactors are divided into:
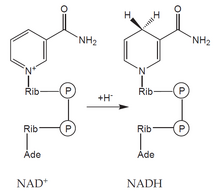
- coenzymes : an organic molecule of a non-protein nature, loosely bound to the apoenzyme molecule - it can be separated from it (e.g. NAD + , NADP + ).
- prosthetic groups : an organic molecule of a non-protein nature, tightly bound to the apoenzyme molecule (e.g. heme , FAD ).
See the Enzyme Cofactors page for more detailed information .
Multienzyme complex[edit | edit source]
An enzyme can be made up of a different number of peptide chains. Each chain can contain multiple domains (with the same or different enzyme specificity). If the enzyme contains multiple chains (quaternary structure), we refer to it as a multienzyme complex . Individual subunits usually have different specificities and are non-covalently linked to each other. An example of a multienzyme complex is fatty acid synthase, which catalyzes the synthesis of higher fatty acids in cells.
Zymogens[edit | edit source]
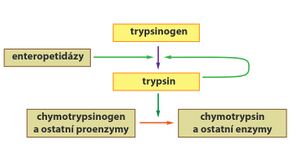
Some enzymes (e.g. digestive) are created and secreted in their inactive form as so-called zymogens (proenzymes) . The reason is the protection of synthesizing cells from being split by the effect of active forms of enzymes. Zymogens are activated only at the point where their activity is required. Activation can, for example, take place as so-called partial proteolysis, during which a precisely defined part of the proenzyme molecule is cleaved off.
Here are two examples of this process:
- The chief cells of the gastric mucosa secrete the proenzyme pepsinogen . The HCl present in the gastric juice aids in the autoactivation of pepsinogen to active pepsin. The reaction also takes place autocatalytically, when already formed pepsin molecules participate in the splitting of pepsinogen .
- Like pepsin, trypsin is synthesized in the pancreas as inactive trypsinogen. In the small intestine, the hexapeptide is subsequently cleaved using the enzyme enteropeptidase (formed by the cells of the intestinal mucosa) to form active trypsin.
Isoenzymes and enzyme isoforms[edit | edit source]
In the organism, there are enzymes called isoenzymes , which catalyze the same reaction, but differ from each other in their physicochemical properties (different affinity to the substrate, K M , sensitivity to inhibitors) and also in their occurrence in tissues . These genetically conditioned differences (a different sequence of DNA nucleotides ), for example, allow a certain regulation of the conditions under which the given reaction will take place in different tissues.
Isoenzymes catalyzing the conversion of glucose to glucose-6-phosphate (phosphorylation of glucose) are illustrative examples - glucokinase (found in hepatocytes and β-cells of the pancreas) and hexokinase (localized in other cells of the body). Glucokinase shows a lower affinity for its substrate – glucose (this is expressed by the so-called Km , for glucokinase it is approximately 10 mmol/l). This means that the enzyme-catalyzed reaction takes place if the blood glucose level reaches a sufficient level (usually after a meal). With normal glycemia (between meals), glucokinase is not very active. The liver thus leaves enough glucose for other tissues that contain hexokinase with a KM valuearound 0.1 mmol/l.
More detailed information can be found on the Embden-Meyerhof-Parnassus track page .
In addition to isoenzymes, enzyme isoforms are also found in the body. These multiple forms of enzymes come from the same gene (same sequence of DNA nucleotides), but differ in different post-translational modifications or alternative splicing . As a result, these enzymes can also catalyze different reactions.
Mechanism of action of enzymes and factors influencing enzyme activity[edit | edit source]
Mechanism of action of enzymes[edit | edit source]
Enzymes , like other catalysts, work on the principle of reducing the activation energy .
For more detailed information, see What Powers Our Cells .
During the first step, the enzyme-substrate (ES) complex is formed. This reaction is typically very fast and reversible. Subsequently, the substrate is transformed into a product under catalysis by the enzyme. The ES complex thus forms an enzyme-product complex (EP), which disintegrates to release the product. This reaction is slow and irreversible.
The reaction is thus divided into several successive steps , in which one or several transition states ES ( transition states ) arise. The activation energy required for the formation of each intermediate and the subsequent conversion of ES to EP is lower than for the direct conversion of substrate to product, even though the overall ΔG of both reactions is the same.
Interaction of substrate and enzyme[edit | edit source]
The substrate interacts with the enzyme molecule in a region called the active site (center). It is formed as follows:
- Enzyme binding site
- The enzyme binding site is a spatially defined, small part of the enzyme molecule containing precisely distributed functional groups (−SH, −OH, acidic and basic amino acids ), whose position corresponds to the structure of the substrate. Non-bonding interactions (H-bridges, electrostatic and hydrophobic interactions, van der Waals forces) are involved in the binding between the enzyme and the substrate . Covalent bonds occur only exceptionally.
- A catalytic site
- The catalytic site contains other groups responsible for the catalytic activity of the enzyme. These groups often come from the cofactor molecule . However, the exact distinction between the binding and catalytic sites is usually problematic.
In addition to the active site, there may also be allosteric sites on the surface of the enzyme , enabling the regulation of enzyme activity due to various effectors (inhibitors or activators).
The original model of the interaction between the substrate molecule and the enzyme (later called the lock and key theory ) created by the German chemist Emil Hermann Fischer at the end of the 19th century assumed that the substrate molecule fits exactly into the enzyme molecule . In the 20th century, this model was modified by the American biochemist Daniel Edward Koshland, Jr., who came up with the claim that the substrate can to some extent induce a conformational change in the site to which it binds (or interact with each other). Therefore, the exact "lock and key" shape is achieved only after binding. It turns out that this induced fit theory describes the interactions between the enzyme and the substrate better than the original model .
Enzyme specificity[edit | edit source]
Enzyme specificity limits the range of action of a certain enzyme . We distinguish two types of specificity:
1. Substrate specificity[edit | edit source]
The enzyme acts only on a limited group of substrates and will not catalyze the reaction for other substrates. Depending on the extent, substrate specificity can be:
Absolute:[edit | edit source]
- The enzyme catalyzes the reaction of only one specific substrate , but will no longer catalyze reactions involving, for example, derivatives of this substrate. An example is urease catalyzing the reaction:
- urea + H 2 O → CO 2 + 2 NH 3
- However, urease cannot catalyze the hydrolysis of methylurea or thiourea.
Group:[edit | edit source]
- This is a more common form of specificity. An enzyme catalyzes the reactions of several similar substrates (typically containing the same functional groups). The affinity for each substrate can be different ( K M therefore differs for individual substrates). An example is carboxypeptidase B , which hydrolyzes peptides from their carboxy-terminus. It preferentially cleaves peptide bonds containing charged amino acids ( arginine , lysine ).
2. Reaction (effect) specificity[edit | edit source]
An enzyme generally catalyzes one type of reaction . An example can be lipases – enzymes that hydrolyze lipids .
Many enzymes act stereospecifically . They attack only certain configurational isomers of the substrates (for example only the L - or only the D - form) probably due to the necessity of binding the substrate to at least three specific sites of the active center of the enzyme (which is a chiral compound) - the opposite stereoisomer does not bind.
Factors affecting enzyme activity and kinetics of single-substrate reactions[edit | edit source]
Substrate concentration[edit | edit source]
Research into the enzyme kinetics of single-substrate reactions was carried out by Leonor Michaelis and Maud Leonora Mentenová . In this chapter, the laws of general kinetics are applied.
See the Kinetics page for more detailed information .
For catalyzed single-substrate reactions, we assume that they proceed in two steps:
(1) {| align = right |(1) |}
where k cat is the rate constant of the catalyzed reaction. It is also called the turnover number and indicates the number of substrate molecules converted by one enzyme molecule per unit of time (most often in one second). Its values range from 4.10^7 (for catalase) to 0.5 (for lysozyme).
- The maximum rate of a catalyzed reaction
It can be expressed as:
(2)
[math]\displaystyle{ v_{max} = k_{cat} \cdot [\mbox{E}]_t }[/math]
where [E] t is the total enzyme concentration.
- Michaelis and Menten equations
The equation describes how the initial velocity at 0 depends on the rate constant k cat as well as where the equilibrium ES is located:
(3)
[math]\displaystyle{ v_0 = \frac{\Delta [\mbox{P}]}{\Delta t} = \frac{k_{cat} \cdot [\mbox{E}]_t \cdot [\mbox{S}]}{K_M + [\mbox{S}]} }[/math]
We assume that the amount of the enzyme does not change (and its concentration is significantly lower than the concentration of the substrate) and also the presence of a certain pseudo-steady state in which the concentration of the ES complex changes much more slowly than the concentrations of S and P.
By substituting equation (2), we can rewrite equation (3) into the form:
[math]\displaystyle{ v_0 = \frac{v_{max} \cdot [\mbox{S}]}{K_M + [\mbox{S}]} }[/math]
K M ( Michael's constant ) is experimentally defined as the substrate concentration at which the rate of the enzyme-catalyzed reaction is equal to half of the maximum rate (it therefore expresses the affinity of the substrate and the enzyme). The lower the K M value , the higher the affinity (a lower [S] is enough to saturate the enzyme by half).
[math]\displaystyle{ \underline{K_M = [\mbox{S}]:} }[/math]
[math]\displaystyle{ v = \frac{v_{max}}{2} }[/math]
The graph shows that the dependence of the rate of the enzyme-catalyzed reaction on the concentration of the substrate has the shape of a hyperbola . If the substrate concentration is small, the rate change curve approximately corresponds to 1st order kinetics (linear increase). But this almost linear increase cannot continue indefinitely due to the limited number of binding sites on the enzyme molecules. At a sufficiently high concentration of the substrate, after saturation of the binding sites of all available enzyme molecules, the maximum speed ( v max ) is thus achieved. The speed does not increase further and remains approximately constant. This part of the curve corresponds to 0th order kinetics (independent of concentration).
K M can then be determined graphically from the hyperbola, but in practice this determination is not used very often. More accurate is the determination using the inverted values of 1/ v and 1/[S] – the so-called double reciprocal plotting according to Lineweaver and Burke. From equation (4) we then get:
(5)
[math]\displaystyle{ \frac{1}{v} = \frac{K_M + [\mbox{S}]}{v_{max} \cdot [\mbox{S}]} }[/math]
We can also write the equation in the form corresponding to the equation of the line y = a · x + b
(6)
[math]\displaystyle{ \frac{1}{v} = \frac{K_M}{v_{max} \cdot [\mbox{S}]} + \frac{1}{v_{max}} }[/math]
Temperature[edit | edit source]
Due to their protein nature , enzymes can be denatured due to high temperature (conformational changes occur, with subsequent destruction of the active site). This usually occurs at temperatures around 55-60°C. The rate gradually decreases until the course of the respective reaction stops completely .
Low temperatures also reduce the activity of enzymes and thus the speed of reactions catalyzed by them. The enzyme has maximum activity at a temperature known as the temperature optimum (for most human enzymes, this is a temperature of around 37 °C).
pH (concentration of H + )[edit | edit source]
Similar to changes in temperature, enzymes are also sensitive to fluctuations in pH . Extreme values (both low and high) can again induce denaturation of the enzyme molecule. The concentration of H + also has a significant effect on the ionization of acidic and basic groups not only of the enzyme molecule, but also of the substrate molecule. Analogously to temperature, the pH optimum is considered to be the pH value at which the enzyme has maximum activity.
Nomenclature of enzymes[edit | edit source]
Trivial terminology[edit | edit source]
The originally used term fermenty , which was based on the fact that enzymes are involved in fermentation, is no longer used. Enzymes discovered among the first were usually named after their source or the method by which they were discovered. Thus, their names tend to be unrelated to the mechanism of the reaction they catalyze. Many end with the suffix -in – see pepsin found in the digestive juice of the stomach (Greek pepsis – digestion) or ptyalin found in saliva (Greek ptyalon – saliva) .
Recommended terminology[edit | edit source]
It introduces a system into the nomenclature and is at the same time simpler than systematic nomenclature, which is why we often use it in everyday practice. We create the name by combining:
- Substrate + -ase (-asa) – for example, amylase (catalyzing the hydrolysis of amylose);
- Type of reaction + -ase – for example, dehydrogenase.
Systematic nomenclature[edit | edit source]
Systematic nomenclature was introduced by IUB ( International Union of Biochemistry ). Each enzyme has its own EC ( Enzyme Commission ) number consisting of four digits – xxxx The first indicates one of the six main enzyme classes, the other two a subgroup and a subgroup. The last one indicates the order of the enzyme in the subgroup (and thus completely characterizes the given enzyme). We recognize these seven main classes of enzymes :
- oxidoreductases,
- transferases,
- hydrolases,
- lyases (synthases),
- isomerase,
- ligases (synthetases),
- translocases.
Oxidoreductases[edit | edit source]
Oxidoreductases catalyze reactions in which one component is oxidized and another is reduced. They often use cofactors – for example NAD + , NADP + , FAD or heme . Oxidoreductases include:
- oxidases, peroxidases,
- oxygenases – introduce oxygen into the substrate molecule , either in the form of -OH (monooxygenase or hydroxylase) or as O 2 (dioxygenase),
- dehydrogenases – oxidize the substrate by removing H-atoms, their name is abbreviated as DH (e.g. lactate dehydrogenase – LDH, alcohol dehydrogenase – ADH),
- desaturases.
Transferases[edit | edit source]
Transferases are involved in the transfer of various groups (amino-, acyl-, methyl-, glycosyl-, phosphoryl-, ...). Examples of transferases are:
- transaminases (aminotransferases) – transfer the -NH 2 group,
- kinases (phosphotransferases) – transfer the phosphate group from ATP or other nucleoside triphosphates,
- transaldolase, transketolase.
Hydrolases[edit | edit source]
They catalyze hydrolytic reactions (the splitting of bonds in molecules by means of a water molecule). Hydrolases include:
- lipases, phospholipases,
- proteases, peptidases (pepsin, trypsin),
- esterases,
- phosphatases.
Lyases (synthases)[edit | edit source]
They catalyze the removal of a certain group from the substrate without hydrolysis (non-hydrolytic cleavage of e.g. bonds between CC, CN) as well as addition reactions to double bonds and syntheses without consumption of ATP. Examples of lyases are:
- decarboxylase,
- aldolase,
- dehydratase, hydratase.
Isomerases[edit | edit source]
They catalyze changes within one substrate molecule (intramolecular changes). The resulting product is an isomer of the starting substrate. Isomerases include:
- epimerase – changes the position of the -OH group in the molecule,
- mutases – change the position of the phosphate group in the molecule.
Ligases (synthetases)[edit | edit source]
They catalyze synthetic reactions associated with ATP hydrolysis (coupling of exergonic and endergonic reactions). Examples of ligases are:
- carboxylase,
- DNA ligase.
Translocases[edit | edit source]
They ensure the movement of substances across the biological membrane. They enable a specific transfer of atoms and molecules. E.g.:
- TOM complex - ensures the transition of the outer mitochondrial membrane (translocase of the outer mitochondrial membrane)
- ADP-ATP-translocase – catalyzes the antiport of ATP behind ADP on the inner mitochondrial membrane
Enzyme inhibition and its importance in pharmacology[edit | edit source]
Enzyme inhibition[edit | edit source]
There are many substances capable of influencing enzyme function in the sense of increasing ( activators ) or decreasing ( inhibitors ) its activity. The inhibition of enzyme activity is one of the most important regulatory mechanisms in living systems. The action of many drugs is also based on the inhibition of specific enzymes of metabolic pathways. Therefore, it is important to know the inhibitory mechanism of their action, which affects, for example, the possibilities of neutralizing their effect. Based on the reversibility of the effect, there are two main forms of inhibition:
- Reversible inhibition,
- Irreversible inhibition.
Reversible inhibition[edit | edit source]
Reversible inhibition can be suppressed. The inhibitor binds non-covalently ( by weak chemical bonds ) either to the active site of the enzyme or outside it. The effect of the inhibitor can be removed, for example, by increasing the supply of the substrate or by dialysis .
- Competitive inhibition
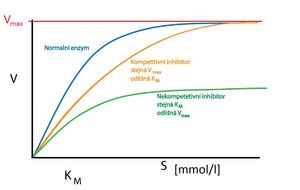
- A competitive inhibitor competes with a substrate molecule for the enzyme's active site. These are often substances structurally similar to the substrate molecule, but incapable of undergoing an enzyme-catalyzed reaction – the inhibitor just binds to the enzyme. By increasing the concentration of the substrate, the inhibition can be suppressed by displacing the inhibitor from the active site.
- A competitive inhibitor does not affect max , it only delays its achievement (the inhibitor must be displaced by an increased concentration of the substrate). Thus, K M increases (seemingly, the affinity of the enzyme for the substrate is reduced).Graph of dependence in max on substrate concentration
- Noncompetitive inhibition
- In noncompetitive inhibition, the inhibitor binds outside the substrate binding site. This location is sometimes referred to as the modulation location. By binding, it changes the conformation of the enzyme in such a way that it also affects the conformation of the active site. This prevents binding of the substrate . Inhibition cannot be suppressed by increasing the concentration of the substrate, because the substrate has no tendency to bind to the modulatory site (so there is no fight - competition for the binding site). This inhibition can only be reversed by removing the inhibitor (e.g. dialysis).
- Since none of the enzyme-inhibitor complexes (or even enzyme-inhibitor-substrate) is catalytically active, the total amount of enzyme available for the substrate is reduced. This will result in a reduction in max reaction. K M does not change in this case .
- Acompetitive inhibition
- This is inhibition in which the inhibitor binds only to the enzyme-substrate complex . This creates a ternary enzyme-inhibitor-substrate complex.
- There is a decrease in max (occupied complexes are enzymatically ineffective) and K M , but their mutual ratio does not change. The inhibitor is very ineffective at low substrate concentration because it does not have enough ES complexes to bind to.
Irreversible inhibition[edit | edit source]
During this inhibition, also referred to as irreversible , a covalent modification of the enzyme molecule occurs . The inhibitor binds covalently to or outside the active site of the enzyme, and therefore it is not possible to remove the inhibition (for example, by dialysis or by increasing the concentration of the substrate).
Examples include heavy metals (Ag + , Hg 2+ ,...) or organophosphates and their derived nerve gases such as sarin and tabun.
Another phenomenon described is inhibition by an excess of substrate . When the concentration of the substrate is too high, there is a fight for the binding site between its molecules. This is reflected in the graph by a slight decrease in max in the area of higher substrate concentration.
Allosteric regulation of enzyme activity[edit | edit source]
Many regulatory enzymes that limit the rate of metabolic pathways (so-called rate-limiting enzymes ) are allosteric enzymes . Allosteric regulation of their activity is one of the most important forms of regulation of metabolic pathways.
In addition to the active site, the allosteric enzyme also has another site on its surface, the so-called allosteric (Greek αλλος – other) , through which it can be affected by modulators (activators or inhibitors). Binding of the allosteric modulator causes a conformational change in the enzyme molecule. This change leads to different affinity for the substrate and other ligands . Most allosteric enzymes are oligomeric (composed of subunits). Binding of a modulator to one subunit will affect the activity of other subunits through a conformational change. We distinguish two basic types of allosteric regulation:
- Homotropic – the modulator is also a substrate for the enzyme. A well-known example is O 2 , which is a homotropic allosteric modulator of hemoglobin .
- Heterotropic – modulator and substrate are different molecules. Following the previous example, CO 2 would be a heterotropic allosteric modulator of hemoglobin.
Allosteric enzymes exhibit sigmoidal kinetics[edit | edit source]
As an example, we will use a reaction affected by a homotropically acting allosteric activator. At low substrate concentrations, the reaction proceeds slowly because only a few enzyme molecules are occupied. If a substrate binds to at least one subunit of the enzyme, it will increase the affinity of the other subunits as well. This will be reflected on the graph by a sharp rise in the reaction rate. The more subunits the enzyme molecule contains, the faster the onset of the effect of increasing the substrate concentration.
The enzyme works on an all-or-nothing basis . Before reaching a certain concentration of the substrate, the reaction almost does not proceed, on the contrary, above the given concentration of the substrate it quickly reaches V max . At this moment, all the binding sites of the enzyme subunits are occupied.
This property of allosteric enzymes is very advantageous in the regulation of metabolic pathways, as it enables the reaction and thus the entire metabolic pathway to be turned on or off quickly.
The use of enzymes in the diagnosis of diseases[edit | edit source]
Determining the activities of various enzymes in body fluids is often used in clinical practice to diagnose the location and extent of tissue damage . In addition to the diagnostic benefit, there is also a significant prognostic benefit, evaluated on the basis of changes in the levels of the relevant enzymes over time.
Enzyme activities are most often measured directly in plasma . It can be:
- Plasma specific enzymes – eg clotting factors .
- Secretory enzymes that enter the blood to varying degrees - e.g. pancreatic amylase or lipase .
- Intracellular enzymes performing various functions in cells – they enter the blood when the cells of the relevant tissue are damaged.
When evaluating measured enzyme levels, it is important to know their:
- Intracellular localization
- Some enzymes are specifically found only in certain compartments of the cell and can thus serve as their markers . An example would be cytochrome oxidase , which is specific for mitochondria . According to the representation of enzymes washed out of the cells, we can also estimate the degree of damage to the given tissue. If only enzymes localized in the cytosol are present in the blood, the impairment is milder than in the case of the occurrence of typically mitochondrial enzymes as well.
- Organ and tissue distribution
- Just as the distribution of enzymes within cellular compartments varies, so does their distribution between individual tissues and organs. Some enzymes are therefore more or less organ specific . Typically, these are mainly various types of isozymes or isoforms of enzymes . Examples are creatine kinase isoenzymes – CK-MB typical for the myocardium and CK-MM for skeletal muscle.
The most frequently determined enzymes[edit | edit source]
Liver[edit | edit source]
- Markers of hepatocyte damage :
- ALT (alanine aminotransferase) – located mainly in the cytoplasm,
- AST (aspartate aminotransferase) - located mainly in mitochondria.
- Since AST is mainly a mitochondrial enzyme, it is released only when liver cells are more severely damaged . The severity of the impairment can therefore be assessed according to the AST/ALT ratio.
- Markers of cholestasis :
- ALP (alkaline phosphatase),
- GGT (γ-glutamyltransferase) – its increased activity is also noted in chronic alcoholic liver disease.
Pancreas[edit | edit source]
- Pancreatic amylase – less specific for pancreatic involvement than pancreatic lipase.
- Pancreatic lipase.
Muscle tissue[edit | edit source]
To distinguish the site of damage, the activities of creatine kinase isoenzymes are measured:
- CK-MM – localized in skeletal muscle;
- CK-MB – located in the myocardium;
- CK-BB – located in the brain.
When diagnosing a myocardial infarction, in addition to CK-MB, the levels are also measured:
- Troponins I and T - currently the most specific cardiac markers, the peak of their concentration occurs about 12 hours after a heart attack.
- Myoglobin - not very specific as a cardiac marker, but it reaches its peak concentration already 2 hours after a heart attack.Cardiomarkers
Bone tissue[edit | edit source]
- ALP (alkaline phosphatase) - localized in osteoblasts - the so-called bone fraction, but it can also come from the liver (the so-called liver fraction), GIT or kidneys,
- ACP (acid phosphatase) – localized in osteoclasts, can also come from the prostate.
Lactate dehydrogenase isoenzymes (LD, LDH)[edit | edit source]
- LD-1 – localized in myocardial cells and erythrocytes.
- LD-2 – located in RES.
- LD-3 – located in the lungs.
- LD-4 - located in the kidneys, pancreas and placenta.
- LD-5 – located in the liver and skeletal muscle.
Overview of the most frequently determined enzymes[edit | edit source]
| Determined enzyme | Organ/tissue damage |
|---|---|
| α-amylase (AMS), pancreatic lipase (LPS | Pancreas |
| Alkaline Phosphatase (ALP) | Bone, liver, GIT, kidney, placenta |
| Acid Phosphatase (ACP) | Bone, prostate |
| creatine kinase (CK) | Skeletal muscle, heart |
| Lactate dehydrogenase (LD) | Heart, liver, skeletal muscle, kidney, erythrocytes |
| Alanine aminotransferase (ALT) | Liver |
| Aspartate aminotransferase (AST) | Liver, heart, skeletal muscle |
| Gamma-glutamyltransferase (GGT) | Liver |
Overview of enzyme cofactors[edit | edit source]
Metal ions and trace elements[edit | edit source]
| Cofactor | Examples of enzymes |
|---|---|
| Zn 2+ | Peptidases, alcohol dehydrogenase |
| Mg2 + | ATP- dependent enzymes , phosphohydrolases |
| Mn 2+ | Superoxide dismutase, arginase |
| Fe 2+ / Fe 3+ | Cytochromes, catalase, peroxidases |
| Cu2 + | Cytochrome oxidase, amino oxidase |
| Mo2 + | Xanthine dehydrogenase |
Organic substances[edit | edit source]
Cofactors of oxidoreductases[edit | edit source]
| Cofactor | Default vitamin | Location/Function |
|---|---|---|
| NAD + , NADP + | Nicotinic acid | Respiratory chain, MK synthesis |
| FAD , FMN | Riboflavin (B 2 ) | Respiratory chain |
| Ubiquinone / ubiquinol | Respiratory chain | |
| Hem | Cytochromes |
Transferase cofactors[edit | edit source]
| Cofactor | Default vitamin | Location/Function |
|---|---|---|
| ATP, GTP | Thiamin (B 1 ) | Transfer of a phosphate residue |
| TDP (thiamine diphosphate) | Thiamin (B 1 ) | Transfer of carbon fragments (oxidative decarboxylation) |
| PALP (pyridoxal phosphate) | Pyridoxine (B 6 ) | Transfer of −NH 2 groups (transamination), decarboxylation of amino acids |
| THF (tetrahydrofolate) | Folate (folic acid) | Transfer of one-carbon fragments |
| CoA (coenzyme A) | Pantothenate | Acyl transfer |
| PAPS (phosphoadenosine phosphosulfate) | Sulfate transfer | |
| SAM (S-adenosylmethionine) | Methylation | |
| B 12 -complex | Cobalamin B 12 | Transmission of CH 3 groups |
Cofactors of lyases[edit | edit source]
| Cofactor | Default vitamin | Location/Function |
|---|---|---|
| PALP (pyridoxal phosphate) | Pyridoxine (B 6 ) | Decarboxylation |
Ligase cofactors[edit | edit source]
| Cofactor | Default vitamin | Location/Function |
|---|---|---|
| ATP | ||
| Carboxybiotin | Biotin | CO 2 transfer (carboxylation) |
Links[edit | edit source]
[edit | edit source]
References[edit | edit source]
- Fontana J., Trnka J., Maďa P., Ivák P. et al.: Transformation of substances and energy in the cell. In: Functions of cells and the human body : Multimedia scripts.